- Syllabus
- 1 Introduction
- 2 Data in Biology
- 3 Preliminaries
- 4 R Programming
- 4.1 Before you begin
- 4.2 Introduction
- 4.3 R Syntax Basics
- 4.4 Basic Types of Values
- 4.5 Data Structures
- 4.6 Logical Tests and Comparators
- 4.7 Functions
- 4.8 Iteration
- 4.9 Installing Packages
- 4.10 Saving and Loading R Data
- 4.11 Troubleshooting and Debugging
- 4.12 Coding Style and Conventions
- 4.12.1 Is my code correct?
- 4.12.2 Does my code follow the DRY principle?
- 4.12.3 Did I choose concise but descriptive variable and function names?
- 4.12.4 Did I use indentation and naming conventions consistently throughout my code?
- 4.12.5 Did I write comments, especially when what the code does is not obvious?
- 4.12.6 How easy would it be for someone else to understand my code?
- 4.12.7 Is my code easy to maintain/change?
- 4.12.8 The
stylerpackage
- 5 Data Wrangling
- 6 Data Science
- 7 Data Visualization
- 8 Biology & Bioinformatics
- 8.1 R in Biology
- 8.2 Biological Data Overview
- 8.3 Bioconductor
- 8.4 Microarrays
- 8.5 High Throughput Sequencing
- 8.6 Gene Identifiers
- 8.7 Gene Expression
- 8.7.1 Gene Expression Data in Bioconductor
- 8.7.2 Differential Expression Analysis
- 8.7.3 Microarray Gene Expression Data
- 8.7.4 Differential Expression: Microarrays (limma)
- 8.7.5 RNASeq
- 8.7.6 RNASeq Gene Expression Data
- 8.7.7 Filtering Counts
- 8.7.8 Count Distributions
- 8.7.9 Differential Expression: RNASeq
- 8.8 Gene Set Enrichment Analysis
- 8.9 Biological Networks .
- 9 EngineeRing
- 10 RShiny
- 11 Communicating with R
- 12 Contribution Guide
- Assignments
- Assignment Format
- Starting an Assignment
- Assignment 1
- Assignment 2
- Assignment 3
- Problem Statement
- Learning Objectives
- Skill List
- Background on Microarrays
- Background on Principal Component Analysis
- Marisa et al. Gene Expression Classification of Colon Cancer into Molecular Subtypes: Characterization, Validation, and Prognostic Value. PLoS Medicine, May 2013. PMID: 23700391
- Scaling data using R
scale() - Proportion of variance explained
- Plotting and visualization of PCA
- Hierarchical Clustering and Heatmaps
- References
- Assignment 4
- Assignment 5
- Problem Statement
- Learning Objectives
- Skill List
- DESeq2 Background
- Generating a counts matrix
- Prefiltering Counts matrix
- Median-of-ratios normalization
- DESeq2 preparation
- O’Meara et al. Transcriptional Reversion of Cardiac Myocyte Fate During Mammalian Cardiac Regeneration. Circ Res. Feb 2015. PMID: 25477501l
- 1. Reading and subsetting the data from verse_counts.tsv and sample_metadata.csv
- 2. Running DESeq2
- 3. Annotating results to construct a labeled volcano plot
- 4. Diagnostic plot of the raw p-values for all genes
- 5. Plotting the LogFoldChanges for differentially expressed genes
- The choice of FDR cutoff depends on cost
- 6. Plotting the normalized counts of differentially expressed genes
- 7. Volcano Plot to visualize differential expression results
- 8. Running fgsea vignette
- 9. Plotting the top ten positive NES and top ten negative NES pathways
- References
- Assignment 6
- Assignment 7
- Appendix
- A Class Outlines
7.9 Network visualization
The most commonly used package to visualize network is igraph.
library(igraph)First, let’s create a graph with 15 nodes.
names <- c("Opal", "Tourmaline", "Emerald", "Citrine", "Jasper", "Moonstone", "Agate", "Chalcedony", "Crystal", "Obsidian", "Quartz", "Topaz", "Beryl", "Sugilite", "Sapphire")Use sample_gnm() to generate a random graph.
set.seed(10)
samp_g <- sample_gnm(
n = 15, # number of vertices
m = 20, # total number of edges
directed = FALSE # if it's a directed network
)To assign color to vertices, use vertex_attr(). gorder() returns the number of vertices.
vertex_attr(samp_g) <- list(name = names, color = rep("slategray1", gorder(samp_g)))To assign color to edges, use vertex_attr(). gsize() returns the total number of edges. Here I am assigning random colors.
edge_attr(samp_g) <- list(color = sample(
x = c("slateblue1", "lightpink1", "steelblue1", "palegreen3"),
size = gsize(samp_g), replace = TRUE
))In real case, when you want to modify the color of a specific edge, you need to first find which edge you want to modify…
edges(samp_g)## [[1]]
## IGRAPH 7328255 UN-- 15 20 -- Erdos renyi (gnm) graph
## + attr: name (g/c), type (g/c), loops (g/l), m (g/n), name (v/c), color
## | (v/c), color (e/c)
## + edges from 7328255 (vertex names):
## [1] Emerald --Citrine Citrine --Jasper Tourmaline--Moonstone
## [4] Emerald --Chalcedony Emerald --Crystal Opal --Obsidian
## [7] Agate --Obsidian Opal --Quartz Jasper --Quartz
## [10] Agate --Quartz Obsidian --Quartz Obsidian --Topaz
## [13] Tourmaline--Beryl Agate --Beryl Quartz --Beryl
## [16] Quartz --Sugilite Emerald --Sapphire Moonstone --Sapphire
## [19] Agate --Sapphire Sugilite --Sapphire
##
## attr(,"class")
## [1] "igraph.edge"Then modify it. For example, modify the 1st edge, which is “Emerald–Citrine.”
edge_attr(samp_g, name = "color")[1] <- "red"To visualize the network, just use plot()!
plot(samp_g)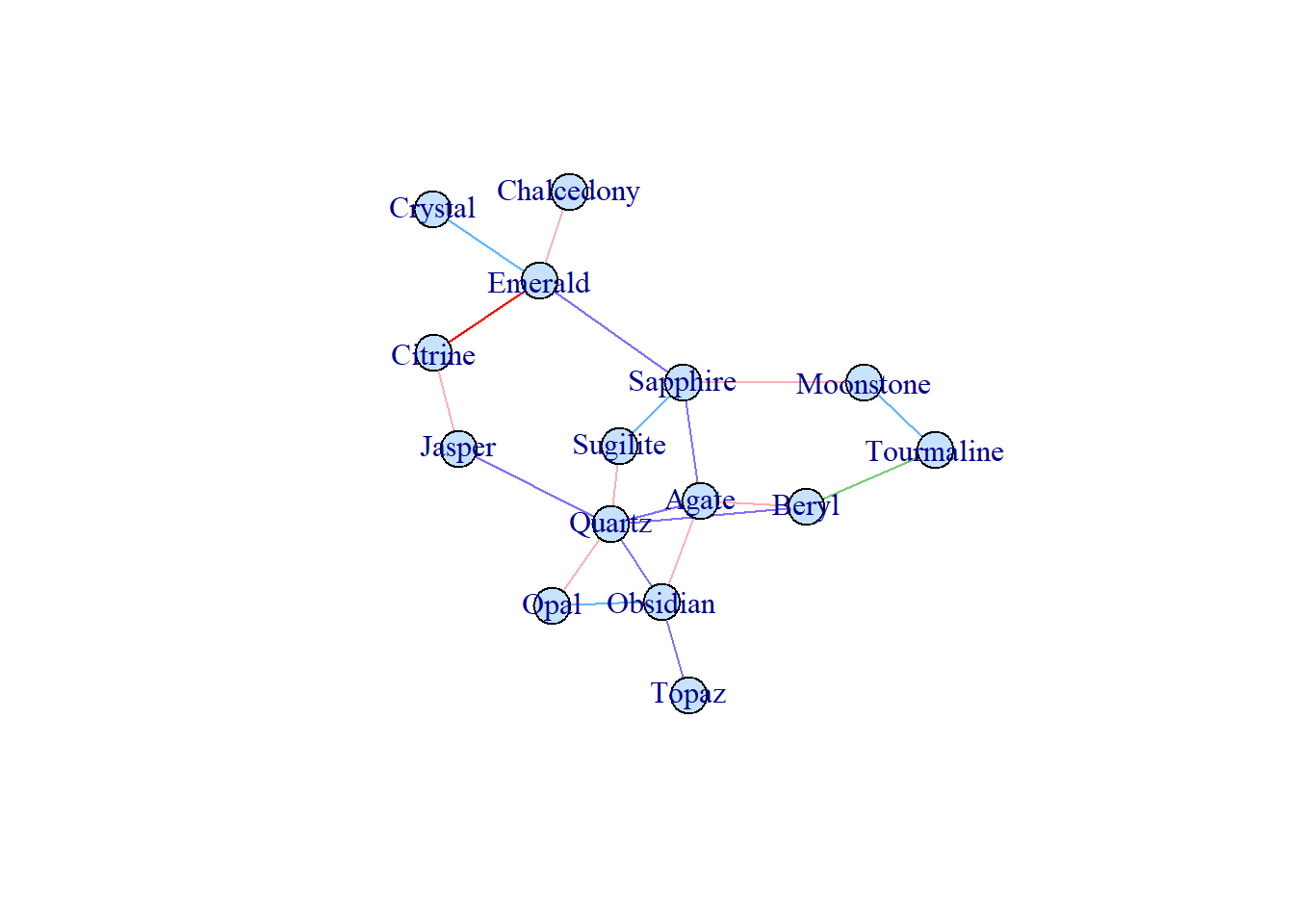
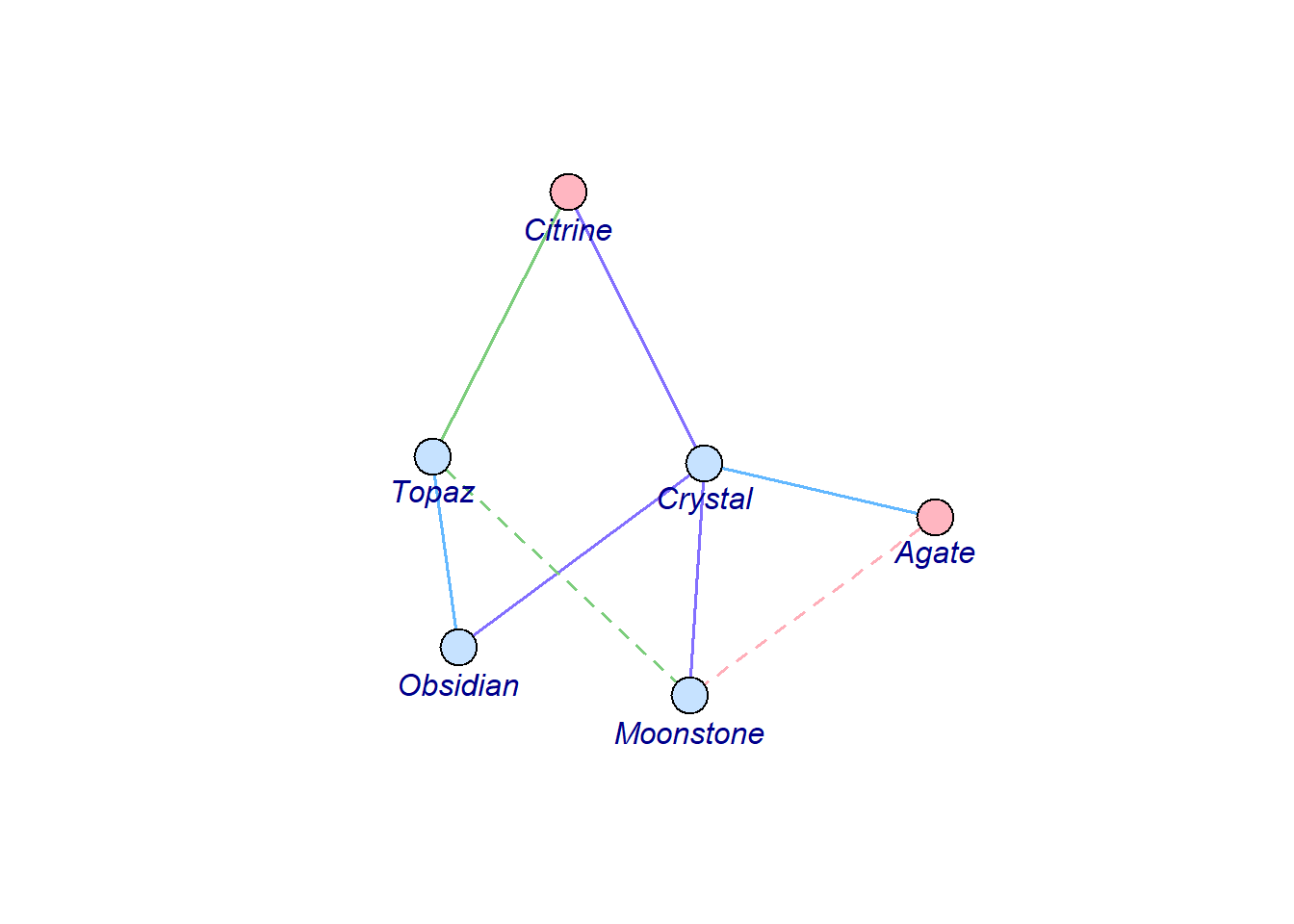
In real-world, we don’t want a random network. Now let’s create our network from scratch.
There are many different ways to create a network. One of the most basic (and easy) ways is using graph_from_data_frame() function in igraph.
1. Create 2 columns in the data frame: from and to. Each row represents an edge, and the entries are vertex names. For instance, as indicated in row 1 below, there is an edge “from Crystal to Moonstone.” In an undirected graph, you can put the two vertices in either way.
2. (optional) Add another column color to represent the color to plot the edge.
3. (optional) You can also add additional information for each edge. For example, add a column weight to specify the weight of this edge. The weight will become useful when you do network analysis later, for example, find the shortest path between two vertices. You can also add other attributes you want, like the “connection_type” here.
set.seed(4)
my_gem_df <- data.frame(
"from" = c("Crystal", "Crystal", "Crystal", "Crystal", "Topaz", "Obsidian", "Agate", "Topaz"),
"to" = c("Moonstone", "Citrine", "Obsidian", "Agate", "Citrine", "Topaz", "Moonstone", "Moonstone"),
"color" = c("slateblue1", "slateblue1", "slateblue1", "steelblue1", "palegreen3", "steelblue1", "lightpink1", "palegreen3"),
"connection_type" = c(rep(1, 6), 2, 2)
)
my_gem_df$weight <- as.numeric(as.factor(my_gem_df$color))
my_gem_df## from to color connection_type weight
## 1 Crystal Moonstone slateblue1 1 3
## 2 Crystal Citrine slateblue1 1 3
## 3 Crystal Obsidian slateblue1 1 3
## 4 Crystal Agate steelblue1 1 4
## 5 Topaz Citrine palegreen3 1 2
## 6 Obsidian Topaz steelblue1 1 4
## 7 Agate Moonstone lightpink1 2 1
## 8 Topaz Moonstone palegreen3 2 2Now create an igraph object from the data frame, and assign vertex color.
g <- graph_from_data_frame(my_gem_df, directed = FALSE)
vertex_attr(g) <- list(
name = c("Crystal", "Topaz", "Obsidian", "Agate", "Moonstone", "Citrine"),
color = c("slategray1", "slategray1", "slategray1", "lightpink", "slategray1", "lightpink")
)Again, simply use plot(g) to plot the network. We can also do some fancy customization to play around with the labels! (Remember the connection_type I specified above in the data frame? In edge.lty, 1 is a solid line, and 2 is a dash line)
set.seed(4)
plot(g,
edge.width = 1.5,
edge.lty = edge_attr(g, "connection_type"),
vertex.label.family = "Trebuchet MS",
vertex.label.font = 3,
vertex.label.cex = 1,
vertex.label.dist = 2,
vertex.label.degree = pi / 2
)
To see the whole list of available arguments available when plotting your network, visit igraph plot manual.
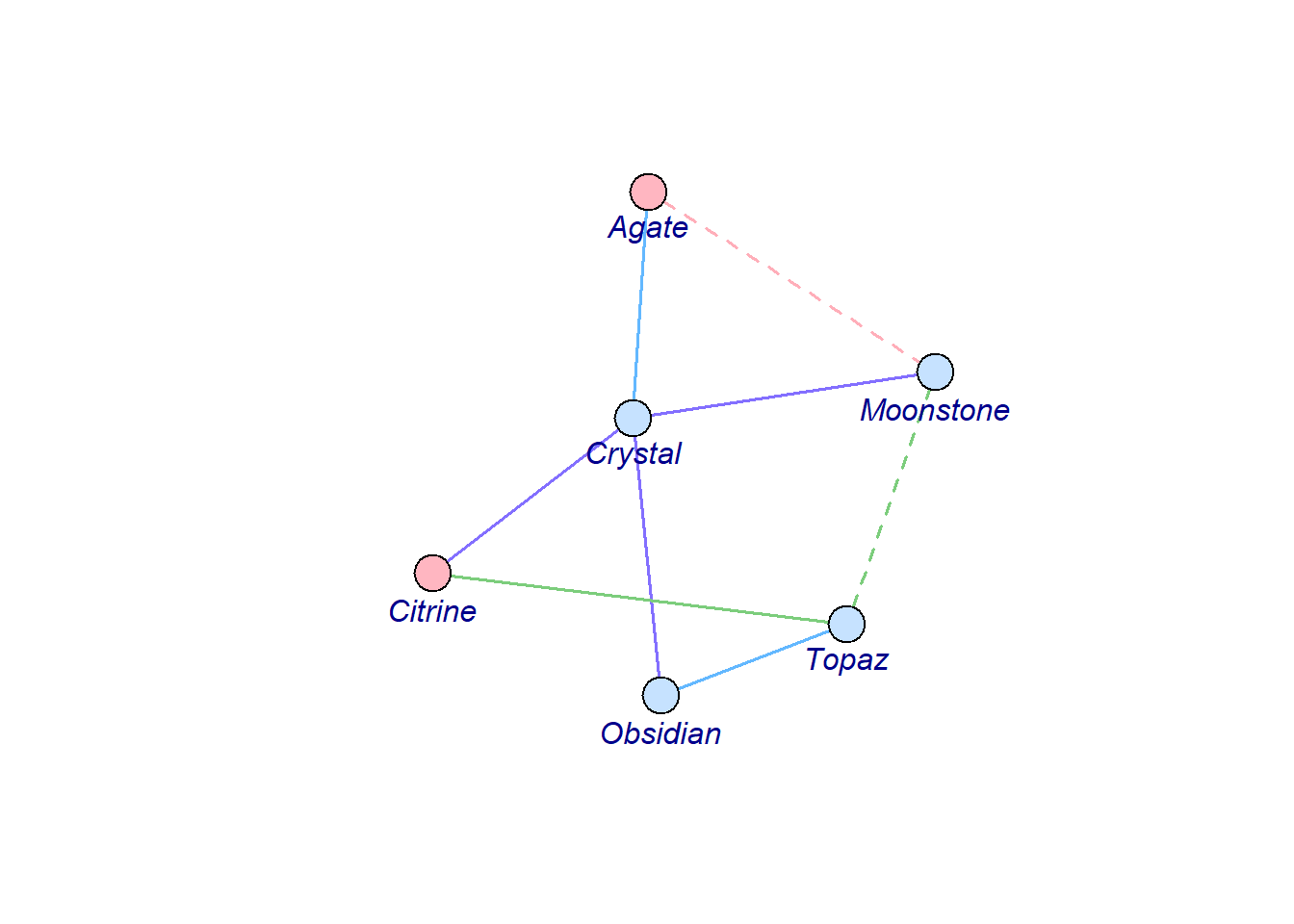
In the case of an directed graph, simply put directed = TRUE when creating the igraph object, and you will see the cute arrows.
g <- graph_from_data_frame(my_gem_df, directed = TRUE)
vertex_attr(g) <- list(
name = c("Crystal", "Topaz", "Obsidian", "Agate", "Moonstone", "Citrine"),
color = c("slategray1", "slategray1", "slategray1", "lightpink", "slategray1", "lightpink")
)
set.seed(4)
plot(g,
edge.width = 1.5,
edge.lty = edge_attr(g, "connection_type"),
edge.arrow.size = 0.8,
vertex.label.family = "Trebuchet MS",
vertex.label.font = 3,
vertex.label.cex = 1,
vertex.label.dist = 2,
vertex.label.degree = pi / 2
)
7.9.1 Interactive network plots
Besides igraph, there are many other packages to create fancier networks. One example is visNetwork. It can create an interactive network, and it’s easy to use! Check example here. It can be implemented in your HTML report knitted from R markdown and R Shiny app interface!
One thing to keep in mind: as you can imagine, since it is interactive, visNetwork requires much more memory and time compared to a static graph. So, you may need to make a choice when you develop your web interface or R markdown HTML report.