- Syllabus
- 1 Introduction
- 2 Data in Biology
- 3 Preliminaries
- 4 R Programming
- 4.1 Before you begin
- 4.2 Introduction
- 4.3 R Syntax Basics
- 4.4 Basic Types of Values
- 4.5 Data Structures
- 4.6 Logical Tests and Comparators
- 4.7 Functions
- 4.8 Iteration
- 4.9 Installing Packages
- 4.10 Saving and Loading R Data
- 4.11 Troubleshooting and Debugging
- 4.12 Coding Style and Conventions
- 4.12.1 Is my code correct?
- 4.12.2 Does my code follow the DRY principle?
- 4.12.3 Did I choose concise but descriptive variable and function names?
- 4.12.4 Did I use indentation and naming conventions consistently throughout my code?
- 4.12.5 Did I write comments, especially when what the code does is not obvious?
- 4.12.6 How easy would it be for someone else to understand my code?
- 4.12.7 Is my code easy to maintain/change?
- 4.12.8 The
stylerpackage
- 5 Data Wrangling
- 6 Data Science
- 7 Data Visualization
- 8 Biology & Bioinformatics
- 8.1 R in Biology
- 8.2 Biological Data Overview
- 8.3 Bioconductor
- 8.4 Microarrays
- 8.5 High Throughput Sequencing
- 8.6 Gene Identifiers
- 8.7 Gene Expression
- 8.7.1 Gene Expression Data in Bioconductor
- 8.7.2 Differential Expression Analysis
- 8.7.3 Microarray Gene Expression Data
- 8.7.4 Differential Expression: Microarrays (limma)
- 8.7.5 RNASeq
- 8.7.6 RNASeq Gene Expression Data
- 8.7.7 Filtering Counts
- 8.7.8 Count Distributions
- 8.7.9 Differential Expression: RNASeq
- 8.8 Gene Set Enrichment Analysis
- 8.9 Biological Networks .
- 9 EngineeRing
- 10 RShiny
- 11 Communicating with R
- 12 Contribution Guide
- Assignments
- Assignment Format
- Starting an Assignment
- Assignment 1
- Assignment 2
- Assignment 3
- Problem Statement
- Learning Objectives
- Skill List
- Background on Microarrays
- Background on Principal Component Analysis
- Marisa et al. Gene Expression Classification of Colon Cancer into Molecular Subtypes: Characterization, Validation, and Prognostic Value. PLoS Medicine, May 2013. PMID: 23700391
- Scaling data using R
scale() - Proportion of variance explained
- Plotting and visualization of PCA
- Hierarchical Clustering and Heatmaps
- References
- Assignment 4
- Assignment 5
- Problem Statement
- Learning Objectives
- Skill List
- DESeq2 Background
- Generating a counts matrix
- Prefiltering Counts matrix
- Median-of-ratios normalization
- DESeq2 preparation
- O’Meara et al. Transcriptional Reversion of Cardiac Myocyte Fate During Mammalian Cardiac Regeneration. Circ Res. Feb 2015. PMID: 25477501l
- 1. Reading and subsetting the data from verse_counts.tsv and sample_metadata.csv
- 2. Running DESeq2
- 3. Annotating results to construct a labeled volcano plot
- 4. Diagnostic plot of the raw p-values for all genes
- 5. Plotting the LogFoldChanges for differentially expressed genes
- The choice of FDR cutoff depends on cost
- 6. Plotting the normalized counts of differentially expressed genes
- 7. Volcano Plot to visualize differential expression results
- 8. Running fgsea vignette
- 9. Plotting the top ten positive NES and top ten negative NES pathways
- References
- Assignment 6
- Assignment 7
- Appendix
- A Class Outlines
2.1 A Brief History of Data in Molecular Biology
Molecular biology became a data science in 1953 when the structure of DNA was determined. Prior to this advance, biochemical assays of biological systems could make general statements about the characteristics and composition of biological macromolecules (e.g. there are two types of nucleic acids - those made of ribose (RNA) and deoxyribose (DNA)) and some quantitative statements about those compositions (e.g. there are roughly equal concentrations of purines - adenine, guanine - and pyrimidines - cytosine, thymine - in any single chromosome). However, once it was shown that each nucleic acid molecule had a specific (and eventually measurable) sequence, this opened the possibility of defining the genetic signature of every living thing on Earth which, in principle, would enable us to understand how life works from its most basic components. A tantalizing prospect, to say the least.
It is perhaps a happy coincidence that our computational and data storage technologies began developing around the same time these molecular biology advances were being made. While mechanical computers had existed for more than a hundred years and arguably longer, the first modern computer, the Atanasoff-Berry Computer (ABC), was invented by John Vincent Atanasoff and Clifford Berry in 1942 at what is now Iowa State University. Over the following decades, the speed, sophistication, and reliability of computing machines increased exponentially, enabling ever larger and faster computations to be performed.
The development of computational capabilities necessitated technologies that stored information that these machines could use, both for instructions to tell the computers what operations they should perform and data they should use to perform them. Until the 1950s, the most commonly available mechanical data storage technologies like writing, phonographic cylinders and disks (a.k.a. records), and punch cards were impractical or unsuitable to create and store the amount of data needed by these computers. A newer technology, magnetic storage, originally proposed in 1888 by Oberlin Smith quickly became the standard way digital computers read and stored information.
With these technological advances, the second half of the 20th century saw rapid advances in our ability to determine and study the properties and function of biological molecular sequences, primarily DNA and RNA (although the first biological sequences scientists determined were proteins composed of amino acids using methods independently invented by Frederick Sanger and Pehr Edman).
In 1970, Pauline Hogeweg and Ben Hesper defined the new discipline of bioinformatics as “the study of informatic processes in biotic systems” (in fact, the original term was proposed in Dutch, Hogeweg and Hesper’s native language). This early form of bioinformatics was a subfield of theoretical biology, studied by those who recognized that biological systems, much like our computer systems, can be viewed as information storage and processing systems themselves.
This broad definition of bioinformatics began narrowing in practice to the study of genetic information as the amount of molecular sequence data we collected grew. By the early 1980s, biological sequence data stored on magnetic tape were being created and studied using new pattern recognition algorithms on computers, which were becoming more widely available at academic and research institutions. At the same time, the idea of determining the complete sequence of the human genome was born, leading to the inception of the The Human Genome Project and the present modern post genomic era.
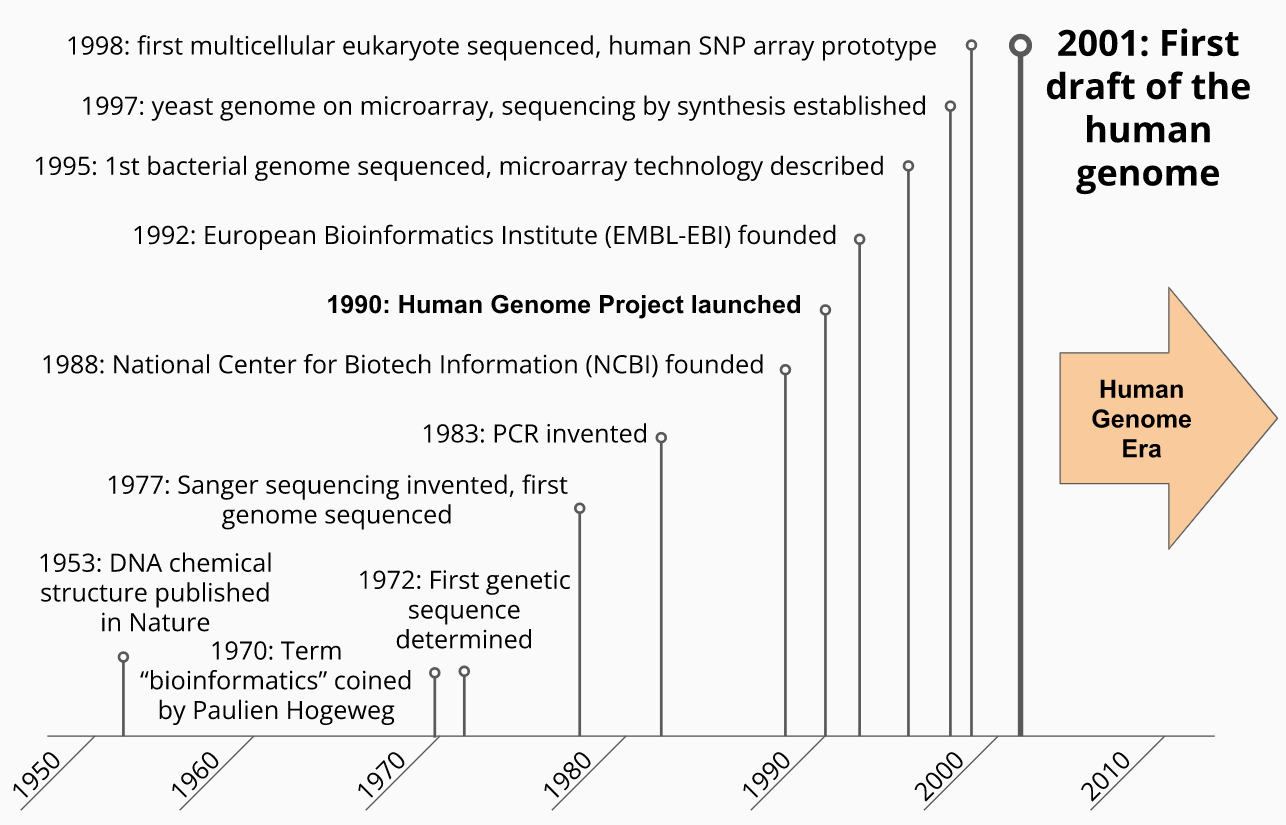
Biological Data Timeline - Setting the Stage