- Syllabus
- 1 Introduction
- 2 Data in Biology
- 3 Preliminaries
- 4 R Programming
- 4.1 Before you begin
- 4.2 Introduction
- 4.3 R Syntax Basics
- 4.4 Basic Types of Values
- 4.5 Data Structures
- 4.6 Logical Tests and Comparators
- 4.7 Functions
- 4.8 Iteration
- 4.9 Installing Packages
- 4.10 Saving and Loading R Data
- 4.11 Troubleshooting and Debugging
- 4.12 Coding Style and Conventions
- 4.12.1 Is my code correct?
- 4.12.2 Does my code follow the DRY principle?
- 4.12.3 Did I choose concise but descriptive variable and function names?
- 4.12.4 Did I use indentation and naming conventions consistently throughout my code?
- 4.12.5 Did I write comments, especially when what the code does is not obvious?
- 4.12.6 How easy would it be for someone else to understand my code?
- 4.12.7 Is my code easy to maintain/change?
- 4.12.8 The
stylerpackage
- 5 Data Wrangling
- 6 Data Science
- 7 Data Visualization
- 8 Biology & Bioinformatics
- 8.1 R in Biology
- 8.2 Biological Data Overview
- 8.3 Bioconductor
- 8.4 Microarrays
- 8.5 High Throughput Sequencing
- 8.6 Gene Identifiers
- 8.7 Gene Expression
- 8.7.1 Gene Expression Data in Bioconductor
- 8.7.2 Differential Expression Analysis
- 8.7.3 Microarray Gene Expression Data
- 8.7.4 Differential Expression: Microarrays (limma)
- 8.7.5 RNASeq
- 8.7.6 RNASeq Gene Expression Data
- 8.7.7 Filtering Counts
- 8.7.8 Count Distributions
- 8.7.9 Differential Expression: RNASeq
- 8.8 Gene Set Enrichment Analysis
- 8.9 Biological Networks .
- 9 EngineeRing
- 10 RShiny
- 11 Communicating with R
- 12 Contribution Guide
- Assignments
- Assignment Format
- Starting an Assignment
- Assignment 1
- Assignment 2
- Assignment 3
- Problem Statement
- Learning Objectives
- Skill List
- Background on Microarrays
- Background on Principal Component Analysis
- Marisa et al. Gene Expression Classification of Colon Cancer into Molecular Subtypes: Characterization, Validation, and Prognostic Value. PLoS Medicine, May 2013. PMID: 23700391
- Scaling data using R
scale() - Proportion of variance explained
- Plotting and visualization of PCA
- Hierarchical Clustering and Heatmaps
- References
- Assignment 4
- Assignment 5
- Problem Statement
- Learning Objectives
- Skill List
- DESeq2 Background
- Generating a counts matrix
- Prefiltering Counts matrix
- Median-of-ratios normalization
- DESeq2 preparation
- O’Meara et al. Transcriptional Reversion of Cardiac Myocyte Fate During Mammalian Cardiac Regeneration. Circ Res. Feb 2015. PMID: 25477501l
- 1. Reading and subsetting the data from verse_counts.tsv and sample_metadata.csv
- 2. Running DESeq2
- 3. Annotating results to construct a labeled volcano plot
- 4. Diagnostic plot of the raw p-values for all genes
- 5. Plotting the LogFoldChanges for differentially expressed genes
- The choice of FDR cutoff depends on cost
- 6. Plotting the normalized counts of differentially expressed genes
- 7. Volcano Plot to visualize differential expression results
- 8. Running fgsea vignette
- 9. Plotting the top ten positive NES and top ten negative NES pathways
- References
- Assignment 6
- Assignment 7
- Appendix
- A Class Outlines
8.4 Microarrays

A Microarray Device - Thermo Fisher Scientific
Microarrays are devices that measure the relative abundance of thousands of distinct DNA sequences simultaneously. Short (~25 nucleotide) single-stranded DNA molecules called probes are deposited on a small glass slide in a grid of spots, where each spot contains many copies of a probe with identical sequence. The probe sequences are selected a priori based on the purpose of the array. For example, gene expression microarrays have probes that correspond to the coding regions of the genome for a species of interest, while genotyping microarrays use sequences with known variants found in a population of genomes, most often the human population. Microarrays of the same type (e.g. human gene expression) all have the same set of probes. The choice of DNA sequence probes therefore determines what the microarray measures and how to interpret the data. The design of a microarray is illustrated in the following figure.
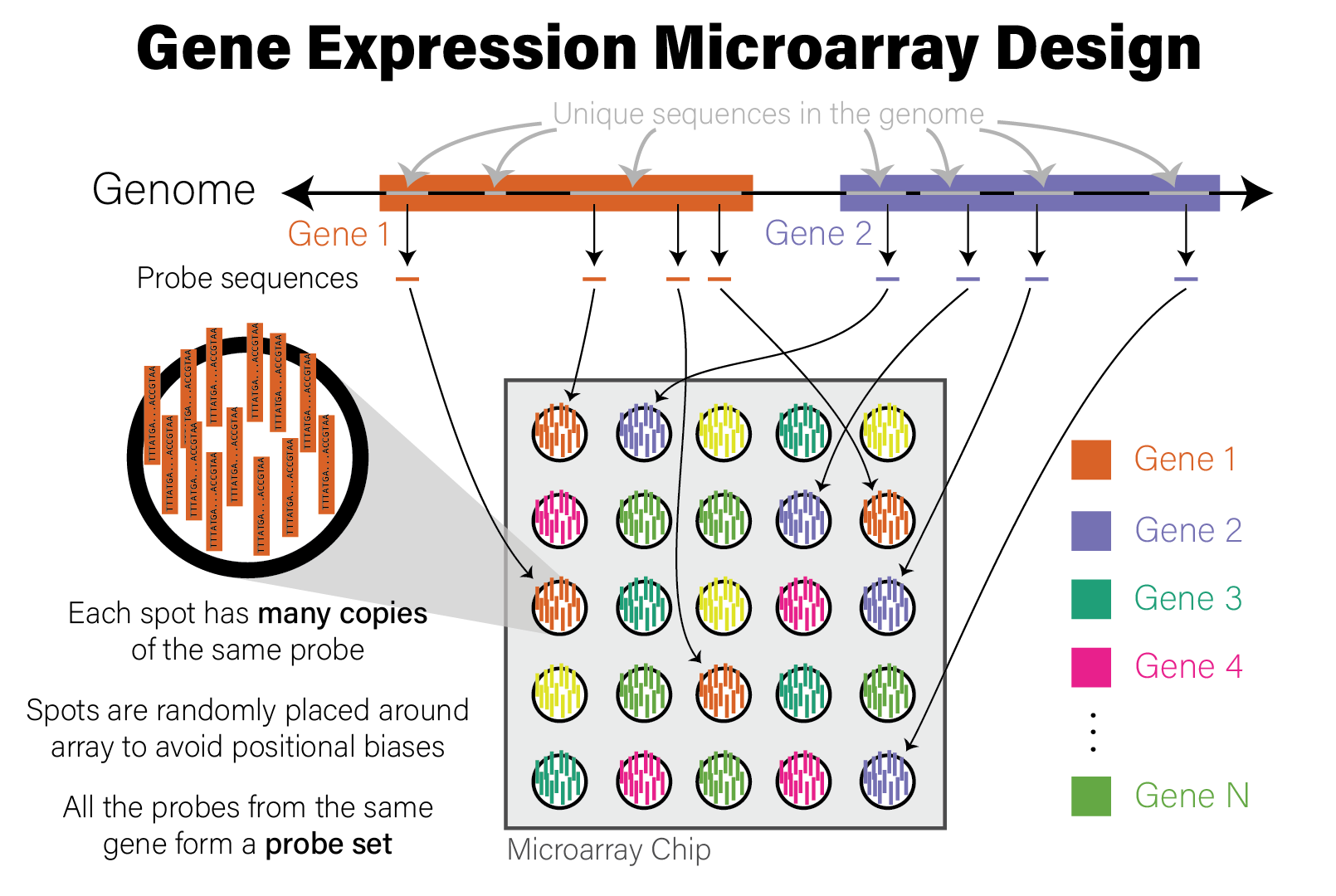
Illustration of Microarray Design
A microarray device generates data by applying a specially prepared DNA sample to it; the sample usually corresponds to a single biological specimen, e.g. an individual patient. The preparation method for the sample depends on what is being measured:
- When measuring DNA directly, e.g. genetic variants, the DNA itself is biochemically extracted
- When measuring gene expression via RNA abundance, RNA is first extracted and then reverse transcribed to complementary DNA (cDNA)
In either case, the molecules that will be applied to the microarray are DNA molecules. After extraction and preparation, the DNA molecules are then randomly cut up into shorter molecules (i.e. sheared) and each molecule has a molecular tag biochemically ligated to it that will emit fluorescence when excited by a specific wavelength of light. This tagged DNA sample is then washed over the microarray chip, where DNA molecules that share sequence complementarity with the probes on the array pair together. After this treatment, the microarray is washed to remove DNA molecules that did not have a match on the array, leaving only those molecules with a sequence match that remain bound to probes.
The microarray chip is then loaded into a scanning device, where a laser with a specific wavelength of light is then shone onto the array, causing the spots with tagged DNA molecules associated with probes to fluoresce, and other spots remain dark. A high resolution image is taken of the fluorescent array, and the image is analyzed to map the intensity of the light on each spot to a numeric value proportional to its intensity. The reason for this is that, since each spot has many individual probe molecules contained within it, the more copies of the corresponding DNA molecule were in the sample, the more light the spot emits. In this way, the relative abundance of all probes on the array are measured simultaneously. The process of generating microarray data from a sample is illustrated in the following figure.
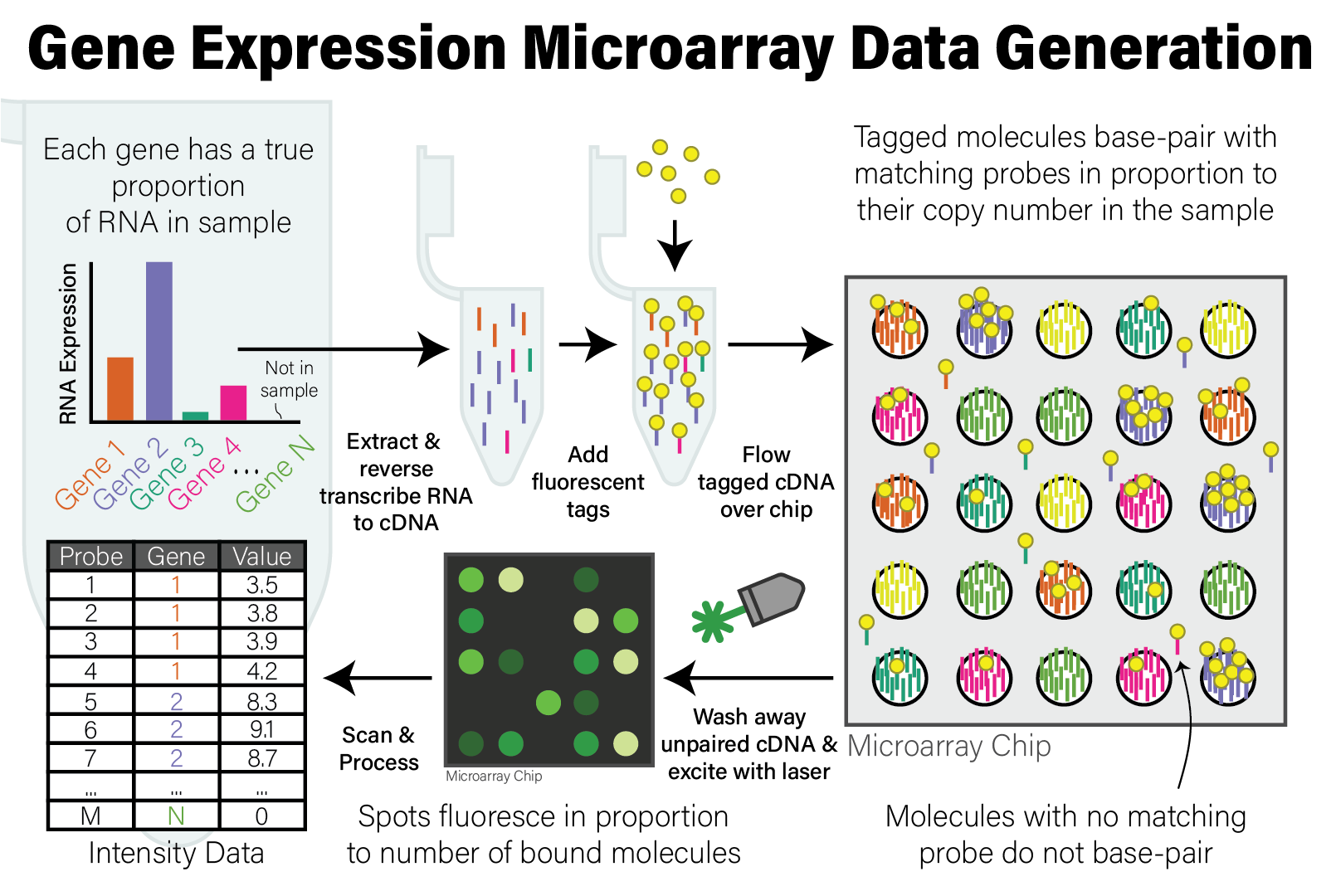
Illustration of Microarray Data Generation Process
After the microarray has been scanned, the relative copy number of the DNA in the sample matching the probes on the microarray are expressed as the intensity of fluorescence of each probe. The raw intensity data from the scan has been processed and analyzed by the scanner software to account for technical biases and artefacts of the scanning instrument and data generation process. The data from a single scan is processed and stored in a file in CEL format, a proprietary data format that stores the raw probe intensity data that can be loaded for downstream analysis.