5 R Programming
5.1 Before you begin
If you have not done so already, be sure to follow the R Language, RStudio, and The R Script, and The Scripting Workflow sections before working through this chapter. The guidance in these sections will set you up for success!
5.2 Introduction
As with other subjects covered so far, the basic syntax of R is covered very
well in other free online materials. Some of those excellent resources are
linked at the end of this section, but a brief overview of the concepts and
syntax are covered here. The code examples below can be written into a script
and evaluated as described above or entered on the R Console directly and run by
pressing Enter.
5.3 R Syntax Basics
At its core, R (like all programming languages) is basically a fancy calculator. The syntax of most basic arithmetic operations in R should be familiar to you:
1 + 2 # addition
[1] 3
3 - 2 # subtraction
[1] 1
4 * 2 # multiplication
[1] 8
4 / 2 # division
[1] 2
1.234 + 2.345 - 3.5*4.9 # numbers can have decimals
[1] -13.571
1.234 + (2.345 - 3.5)*4.9 # expressions can contain parentheses
[1] -4.4255
2**2 # exponentiation
[1] 4
4**(1/2) # square root
[1] 2
9**(1/3) # cube root
[1] 3The [1] lines above are the output given by R when the preceding expression
is executed. Any portion of a line starting with a # is a comment and ignored
by R.
R also supports storing values into symbolic placeholders called variables, or
objects. An expression like those above can be assigned into a variable with a
name using the <- operator:
new_var <- 1 + 2Variables that have been assigned a value can be placed in subsequent expressions anywhere where their value is evaluated:
The correct way to assign a value to a variable in R is with the <- syntax,
unlike many other programming languages which use =. However, although the =
assignment syntax does work in R:
new_var = 2 # works, but is not common convention!this is considered bad practice and may cause confusion later. You should always
use the <- syntax when assigning values to variables!
In R, the period . does not have a special meaning like it does in many other
languages like python, C, javascript, etc. Therefore, new.var is a valid
variable name just like new_var, even though it may look strange to those
familiar with these other languages. While including . in your R variable
names is valid, the results that you will use in programs written in other
languages that do have a meaning for this character. Therefore, it is good
practice to avoid using . characters in your variable names to reduce the
chances of conflicts later.
5.4 Basic Types of Values
The most common type of value in R is the number, e.g. 1.0 or 1e-5 for
\(10^{-5}\). For most practical purposes, R does not distinguish between numbers
with fractional parts (e.g. 1.123) and integers (e.g. 1); a number is a
number. In addition to numbers, there are some other types of values that are
special in R:
-
logical or boolean values -
TRUEorFALSE. Internally, R storesTRUEas the number1andFALSEas the number0. Generally, R interprets non-zero numbers asTRUEand0asFALSE, but it is good practice to supply the tokensTRUEorFALSEwhen an argument expects a logical value. -
missing values -
NA.NAis a special value that indicates a value is missing. -
missing vectors -
NULL. Similar toNA,NULLindicates that a vector, rather than a value, is missing. Vectors will be described in the next section on data strutures. - factors - Factors are a complex type used in statistical models and are covered in greater detail later
-
infinity -
Infand-Inf. These values encode what R understands to be positive or negative infinity, or any number divided by0. -
impossible values -
NaN. This value corresponds to the mathematically ‘impossible’ or undefined value of0/0. -
character data -
"value". R can store character data in the form of strings. Note R does not interpret string values by default, so"1"and1are distinct. - dates and times - R has a basic type to store dates and times (together termed a datetime, which includes both components). Internally, R stores datetimes as the fractional number of days since January 1, 1970, using negative numbers for earlier dates.
-
complex numbers - R can store complex numbers using the
complexfunction.
Unsurprisingly, R cannot perform computations on NA, NaN, or Inf values.
Each of these values have an ‘infectious’ quality to them, where if they are
mixed in with other values, the result of the computation reverts to the first
of these values encountered:
# this how to create a vector of 4 values in R
x <- c(1,2,3,NA)
mean(x) # compute the mean of values that includes NA
[1] NA
mean(x,na.rm=TRUE) # remove NA values prior to computing mean
[1] 2
mean(c(1,2,3,NaN))
[1] NaN
mean(c(NA,NaN,1))
[1] NAIf your code produces values that are not numbers as you expect, this suggests there are one of these values in your input, and need to be handled explicitly.
5.5 Data Structures
5.5.1 Vectors
Data structures in R (and other languages) are ways of storing and organizing more than one value together. The most basic data structure in R is a one dimensional sequence of values called a vector:
The vector in R has a special property that all values contained in the vector must have the same type, from the list described above. When constructing a vector, R will coerce values to the most general type if it encounters values of different types:
c(1,2,"3")
[1] "1" "2" "3"
c(1,2,TRUE,FALSE)
[1] 1 2 1 0
c(1,2,NA) # note missing values stay missing
[1] 1 2 NA
c("1",2,NA,NaN) # NA stays, NaN is cast to a character type
[1] "1" "2" NA "NaN"In addition to having a single type, vectors also have a length, which is defined as the number of elements in the vector:
Internally, R is much more efficient at operating on vectors than individual elements separately. With numeric vectors, you can perform arithmetic operations on vectors of compatible size just as easily as individual values:
c(1,2) + c(3,4)
[1] 4 6
c(1,2) * c(3,4)
[1] 3 8
c(1,2) * c(3,4,5) # operating on vectors of different lengths raises warning, but still works
[1] 3 8 5
Warning message:
In c(1, 2) * c(3, 4, 5) :
longer object length is not a multiple of shorter object lengthIn the example above, we multiplied a vector of length 2 with a vector of length 3:
c(1,2) * c(3,4,5) # operating on vectors of different lengths raises warning, but still works
[1] 3 8 5
Warning message:
In c(1, 2) * c(3, 4, 5) :
longer object length is not a multiple of shorter object lengthRather than raise an error and aborting, R merely emits a warning message about the vectors not having divisible lengths. So how did R decide the third value should be 5? Because R cycles through each vector and multiplies the values element-wise until the longest vector has had an operation performed on all its values:
c(1,2) * c(3,4,5) # yields: 1*3 2*4 1*5
[1] 3 8 5
Warning message:
In c(1, 2) * c(3, 4, 5) :
longer object length is not a multiple of shorter object length
c(1,2) * c(3,4,5,6) # yields: 1*3 2*4 1*5 2*6
[1] 3 8 5 12R will sometimes work in ways you don’t expect. Be careful to read warnings and check that your code does what you expect!
5.5.2 Factors
Factors are objects that R uses to handle categorical variables, i.e. variables
that can take one of a distinct set of values for each sample. For example,
a variable indicating whether a subject had a disease or was a control could be
encoded using a factor with values Disease or Control. Consider an example
dataset with six subjects where three are disease and three are control, and
we create a factor from a corresponding variable of character strings using the
factor() function:
case_status <- factor(
c('Disease','Disease','Disease',
'Control','Control','Control'
)
)
case_status
[1] Disease Disease Disease Control Control Control
Levels: Control DiseaseThe factor case_status prints as a vector of labels, either Disease or
Control. The distinct values in the factor are called levels, and this
factor has two: Control and Disease. Internally, a factor is stored as a
vector of integers where each level has the same value:
as.numeric(case_status)
[1] 2 2 2 1 1 1
str(case_status)
Factor w/ 2 levels "Control","Disease": 2 2 2 1 1 1By default, R assigns integers to levels in alphanumeric order; since “Control”
comes lexicographically before “Disease”, the Control level is assigned the
integer 1 and Disease is assigned 2. Each value of the factor corresponds
to these integers, and since Disease came before Control, the numeric values
of the factor are (2, 2, 2, 1, 1, 1). The integer values assigned to each level
allow the factor to be sorted:
Note the order of the factor levels has changed so that controls, which have a value of 1, precede disease, which have a value of 2. The integers assigned to each level can be specified explicitly when creating the factor:
case_status <- factor(
c('Disease','Disease','Disease','Control','Control','Control'),
levels=c('Disease','Control')
)
case_status
[1] Disease Disease Disease Control Control Control
Levels: Control Disease
str(case_status)
Factor w/ 2 levels "Disease","Control": 1 1 1 2 2 2The base R functions for reading in CSV Files load columns with character
values as factors by default (you may turn this off with
stringsAsFactors=FALSE to read.csv()), and in other situations you may have
factors created by other functions that need to have their integer values
changed. This process is called releveling the factor, and may be accomplished
by passing a factor into the factor() function and specifying new levels:
str(case_status)
Factor w/ 2 levels "Disease","Control": 1 1 1 2 2 2
factor(case_status, levels=c("Control","Disease"))
Factor w/ 2 levels "Control","Disease": 2 2 2 1 1 1Controlling the order of levels in a factor is important in a number of situations. One is when specifying the reference category for categorical variables when constructing model matrices to pass to statistical models, the details of which are beyond the scope of this book. A second is when the order of categorical variables when passed to ggplot, which is covered in greater detail in [Reordering 1-D Data Elements] in the Grammar of Graphics chapter. The forcats tidyverse package provides more powerful functions for working with categorical variables stored in factors.
5.5.3 Matrices
A matrix in R is simply the 2 dimensional version of a vector. That is, it is a rectangle of values that all have the same type, e.g. number, character, logical, etc. A matrix may be constructed using the vector notation described above and specifying the number of rows and columns the matrix should have, and Instead of having a length like a vector, it has \(m \times n\) dimensions:
# create a matrix with two rows and three columns containing integers
A <- matrix(c(1,2,3,4,5,6)
nrow = 2, ncol = 3, byrow=1
)
A
[,1] [,2] [,3]
[1,] 1 2 3
[2,] 4 5 6
dim(A) # the dim function prints out the dimensions of the matrix, rows first
[1] 2 3Because a matrix is 2 dimensional, it can be transposed from \(m \times n\) to be
\(n \times m\) using the t() function:
R can store hyper-rectangular data using arrays. arrays are constructed and
operate similarly to matrix, but may have arbitrarily many dimensions. The
following example is a 3-dimensional array with dimension sizes of \(2x2x3\):
5.5.4 Lists and data frames
Vectors and matrices have the special property that all items must be of the same type, e.g. numbers. Lists and data frames are data structures that do not have this requirement. Similar to vectors, lists and data frames are both one dimensional sequences of values, but the values can be of mixed types. For instance, the first item of a list may be a vector of numbers, while the second is a vector of character strings. These are the most flexible data structures in R, and are among the most commonly used.
Lists can be created using the list() function:
my_list <- list(
c(1,2,3),
c("A","B","C","D")
)
my_list
[[1]]
[1] 1 2 3
[[2]]
[1] "A" "B" "C" "D"
my_list[[1]] # access the first item of the list
[1] 1 2 3
my_list[[2]] # access the second item of the list
[1] "A" "B" "C" "D"The arguments passed to list() define the values and their order of the list.
In the above example, the list has two elements: one vector of 3 numbers and one
vector of 4 character strings. Note you can access individual items of the list
using the [[N]] syntax, where N is the 1-based index of the element.
Lists can also be defined and indexed by name:
my_list <- list(
numbers=c(1,2,3),
categories=c("A","B","C","D")
)
my_list
$numbers
[1] 1 2 3
$categories
[1] "A" "B" "C" "D"
my_list$numbers # access the first item of the list
[1] 1 2 3
my_list$categories # access the second item of the list
[1] "A" "B" "C" "D"The elements of the list have been assigned the names numbers and categories
when creating the list, though any valid R identifier names can be used. When
elements are associated with names they can be accessed using the list$name
syntax.
Lists and data frames are the same underlying data structure, however differ in
one important respect: the elements of a data frame must all have the same
length, while the elements of a list do not. You may create a data frame with
the data.frame() function:
my_df <- data.frame( # recall '.' has no special meaning in R
numbers=c(1,2,3),
categories=c("A","B","C","D")
)
Error in data.frame(c(1, 2, 3), c("A", "B", "C", "D")) :
arguments imply differing number of rows: 3, 4
my_df <- data.frame(
numbers=c(1,2,3),
categories=c("A","B","C")
)
my_df
numbers categories
1 1 A
2 2 B
3 3 C
my_df$numbers
[1] 1 2 3
my_df[1] # numeric indexing also works, and returns a subset data frame
numbers
1 1
2 2
3 3
my_df[1]$numbers
[1] 1 2 3
# this syntax is [<row>,<column>], and if either is omitted return all
my_df[,1] # return all rows of the first column as a vector
[1] 1 2 3
my_df$categories
[1] "A" "B" "C"Note the data frame is printed as a matrix with element names as columns and automatically numbered rows. You may access specific elements of a data frame in a number of ways:
5.5.5 Subsetting
Each of the data structures in R are collections of objects. Very often, we would like to select a subset of elements in a collection that have certain properties, for instance numeric values less than a specific threshold or occupy a specific position in the vector (e.g. the first or last element). The process of selecting out a subset of elements in a data structure is called subsetting. R provides many different methods to subset a data structure, where the choice of method depends on the type of data structure (e.g. vector vis a vis a data frame) or the type of subsetting required by a particular use case.
Unlike other languages like C and python, R uses 1-based indexing. This means that the first item in any data structure is referenced with the index 1:
x <- c(3.5, 0.4, 9.1, 7.7)
x[1]
[1] 3.5
x[4]
[1] 7.7
x[0] # this always returns an empty vector
[1] numeric(0)
x[5] # accessing indices larger than the vector length returns NA
[1] NAC and python use 0-based indexing, where the first item in a data structure is referenced with the index 0. For example:
x = [3.5, 0.4, 9.1, 7.7]
print(x[0])
3.5
print(x[3])
7.7
print(x[4]) # accessing indices larger than the list length raises error
Traceback (most recent call last):
File "<stdin>", line 1, in <module>
IndexError: list index out of rangeAs you move between developing code in 0-based languages and R, be sure to remember they use different indexing strategies!
5.5.5.1 Vectors
There are six different ways to subset vectors in R. The following three are the most common:
-
Positive integers: return specific elements by integer index.
-
Negative integers: omit specific elements by their negative integer index.
-
Logical vectors: return specific elements where the indexing vector is
TRUE
To see the other methods of indexing vectors, see the section on subsetting vectors and Hadley Wickham’s book Advanced R.
5.5.5.2 Matrices
Since matrices in R are vectors, they may be subset using the same methods as
vectors. Additionally, because matrices also have a notion of rows and columns,
they may also be subset by pairs of vectors that select either rows or columns.
A matrix x may be subset with the syntax [<row selectors>, <column selectors>].
The selectors may be any of the methods used to subset a vector, as described
above.
x <- matrix(1:9, nrow=3, byrow=TRUE)
x
[,1] [,2] [,3]
[1,] 1 2 3
[2,] 4 5 6
[3,] 7 8 9
x[c(1,2), c(1,2)] # first two rows, first two columns
[,1] [,2]
[1,] 1 2
[2,] 4 5
x[-c(1,3), ] # leaving a selector blank selects all, second row and all columns
[1] 4 5 6
x[, c(2,3)] # all rows included, last two columns
[,1] [,2]
[1,] 2 3
[2,] 5 6
[3,] 8 9
x[-c(1,3), c(2)] # select the second row and second column
[1] 55.5.5.3 Lists
A list can be subset with all the same methods as a vector.
l <- list(3.5, 0.4, c(9.1, 5.8), 7.7))
l[1]
[[1]]
[1] 3.5
l[c(2,3)]
[[1]]
[1] 0.4
[[2]]
[1] 9.1 5.8
l[c(FALSE, TRUE, TRUE, FALSE)]
[[1]]
[1] 0.4
[[2]]
[1] 9.1 5.8Note that when indexing a list with [, the result returned is always another
list; we will discuss this more later.
When a list has names associated with its values, it may also be access by name in addition to the ways described above:
l <- list(a=3.5, b=0.4, c=c(9.1, 5.8), d=7.7)
l["a"]
$a
[1] 3.5
l[c("a","c")]
$a
[1] 3.5
$c
[1] 9.1 5.8
l[c("b")]We will discuss what $ means at the end of this section.
5.5.5.4 Data frames
Data frames may be accessed like both vectors (and therefore lists) and also like matrices, since they are rectangular by construction.
5.5.5.5 [[ and $
The [[ and $ operators are used to access single items of data structures.
Remember from the section on subsetting lists that indexing with [ (i.e. a
single square bracket) always returns a list. When we wish to access an
individual value of a list or data frame and return only that value, we use
[[ when indexing by integer or name:
l <- list(a=3.5, b=0.4, c=c(9.1, 5.8), d=7.7)
l[1] # returns a list with a single value of 3.5
[[1]]
[1] 3.5
l[[1]] # returns 3.5
[1] 3.5
l[[3]] # returns c(9.1, 5.8)
[1] 9.1 5.8
l["a"] # returns list(a=3.5)
$a
[1] 3.5
l$a # returns 3.5
[1] 3.5
l[["a"]] # also returns 3.5
[1] 3.5To see another (ane more complete) treatment of subsetting, see the section on subsetting vectors and Hadley Wickham’s book Advanced R.
5.5.6 Naming Data Structures
Vectors, matrices, lists, and data frames can have names assigned to their
indices. For vectors and lists, these names are one-dimensional vectors of
characters that are assignable and accessible using the names() function:
x <- c(1,2,3)
names(x) # vectors have no names by default
NULL
names(x) <- c("a","b","c")
x
a b c
1 2 3
names(x)
[1] "a" "b" "c"
names(x) <- c("d","e","f")
x
d e f
1 2 3When a list is created with named items, those names are accessible similarly:
l <- list(a=3.5, b=0.4, c=c(9.1, 5.8), d=7.7)
names(l)
[1] "a" "b" "c" "d"
names(l) <- c("f1", "f2", "f3", "f4")
l
$f1
[1] 3.5
$f2
[1] 0.4
$f3
[1] 9.1 5.8
$f4
[1] 7.7As matrices and data frames also have both rows and columns, they can have
rownames and colnames, respectively:
5.6 Logical Tests and Comparators
As mentioned above, R recognizes logical values as a distinct type. R provides all the conventional infix logical operators:
1 == 1 # equality
[1] TRUE
1 != 1 # inequality
[1] FALSE
1 < 2 # less than
[1] TRUE
1 > 2 # greater than
[1] FALSE
1 <= 2 # less than or equal to
[1] TRUE
1 >= 2 # greater than or equal toThese operators also work on vectors, albeit with the same caveats about vector length as noted earlier:
x <- c(1,2,3)
x == 2
[1] FALSE TRUE FALSE
x < 1
[1] FALSE FALSE FALSE
x < 3
[1] TRUE TRUE FALSE
c(1,2) == c(1,3)
[1] TRUE FALSE
c(1,2) != c(1,3)
[1] FALSE TRUE
c(1,2) == c(1,2,3)
[1] TRUE TRUE FALSE
Warning message:
In c(1, 2, 3) == c(1, 2) :
longer object length is not a multiple of shorter object lengthWhen performing logical tests on vectors, the all() function may be used to
return a single boolean value when all results are true:
R also provides many functions of the form is.X where X is some type or
condition (recall that . is not a special character in R):
5.7 Functions
Just as a variable is a symbolic representation of a value, a function is a
symbolic representation of code. In other words, a function allows you to
substitute a short name, e.g. mean, for a set of operations on a given input,
e.g. the sum of a set of numbers divided by the number of numbers. R provides a
very large number of functions for common operations in
its default environment, and more functions are provided by packages you can
install separately.
Encapsulating many lines of code into a function is useful for (at least) five distinct reasons:
- Make your code more concise and readable
- Allow you to avoid writing the same code over and over (i.e. reuse it)
- Allow you to systematically test pieces of your code to make sure they do what you intend
- Allow you to share your code easily with others
- Program using a functional programming style (see note box below)
At its core, R is a functional programming language. The details of what this means are outside the scope of this book, but as the name implies this refers to the language being structured around the use of functions. While this property has technical implications on the structure of the language, a more important consequence is the style of programming it entails. The functional programming style (or paradigm) has many advantages, including generally producing programs that are more concise, predictable, provably correct, and performant. provides a good starting point for learning about functional programming.
In order to do anything useful, a function must generally be able to accept
and execute on different inputs; e.g. the mean function wouldn’t be very
useful if it didn’t accept a value! The terminology used in R and many other
programming languages for this is the function must accept or allow you to
pass it arguments. In R, functions accept arguments using the following
*pattern:
# a basic function signature
function_name(arg1, arg2) # function accepts 2 argumentsHere, arg1 and arg2 are arguments or actual arguments, indicating
this function accepts two arguments. The name of the function (i.e.
function_name) and the pattern of arguments it accepts is called the
function’s signature. Every function has at least one signature, and it is
critical to understand it in order to use the function properly.
In the above example, arg1 and arg2 are positional arguments. This means
the order of arguments is important. Calling the function above like
function_name(arg2, arg1) will produce a different result than with
function_name(arg1, arg2). As we will see shortly, functions have names for
their arguments in their definition that can be used to specify arguments in any
order.
For most functions, some arguments are required. This means that the function will not execute without two arguments provided and will raise an error if you try otherwise:
mean() # compute the arithmetic mean, but of what?
Error in mean.default() : argument "x" is missing, with no defaultHow do you know what arguments a function requires? All functions provided by
base R and many other packages include detailed documentation that can be
accessed directly through RStudio using either the ? or help():
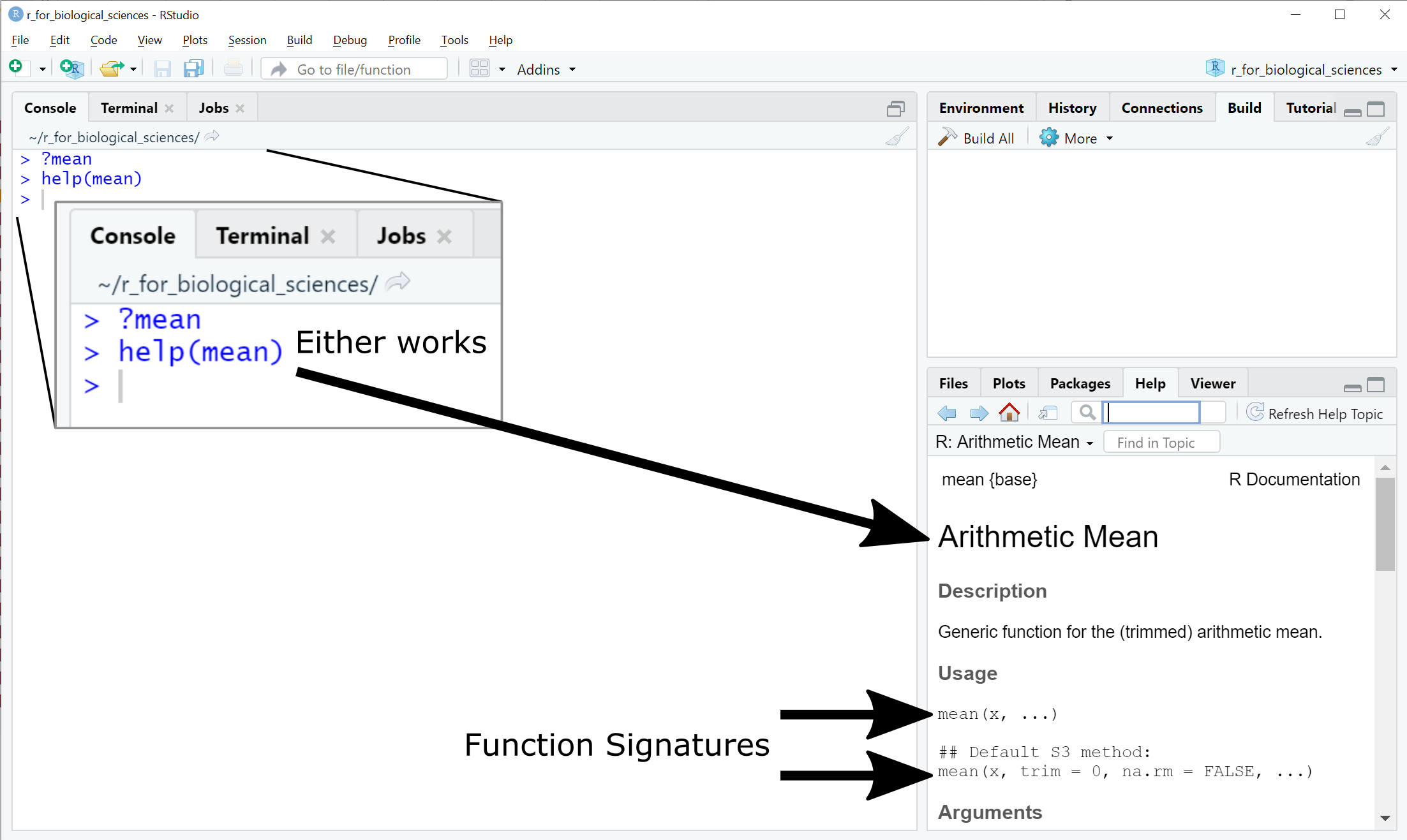
The second signature of the mean function introduces two new types of syntax:
Default argument values - e.g. trim = 0. These are formal arguments that
have a default value if not provided explicitly.
Variable arguments - .... This means the mean function can accept
arguments that are not explicitly listed in the signature. This syntax is called
dynamic dots.
With these definitions, we can now understand the Arguments section of the help documentation:
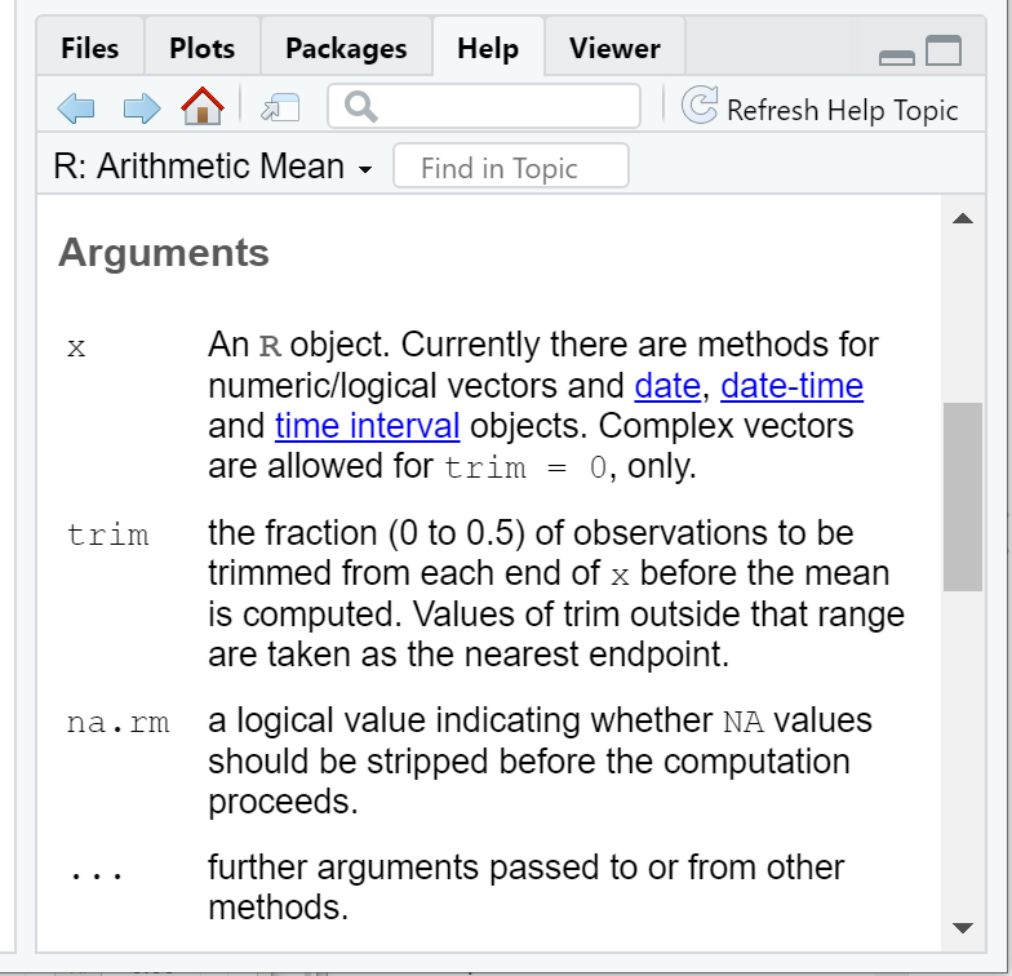
In other words:
-
xis the vector of values (more on this in the next section on data structures) we wish to compute the arithmetic mean for -
trimis a fraction (i.e. a number between 0 and 0.5) that instructs R to remove a portion of the largest and smallest values fromxprior to computing the mean. -
na.rmis a logical value (i.e. eitherTRUEorFALSE) that instructs R to removeNAvalues fromxbefore computing the mean.
All function arguments can be specified by name, regardless of whether there is
a default value or not. For instance, the following two mean calls are
equivalent:
# this generates 100 normally distributed samples with mean 0 and standard deviation 1
my_vals <- rnorm(100,mean=0,sd=1)
mean(my_vals)
[1] -0.05826857
mean(x=my_vals)
[1] -0.05826857To borrow from the Zen of Python, “Explicit is better than implicit.” Being explicit about which variables are being passed as which arguments will almost always make your code easier to read and more likely to do what you intend.
The ... argument catchall can be very dangerous. It allows you to provide
arguments to a function that have no meaning, and R will not raise an error.
Consider the following call to mean:
# this generates 100 normally distributed samples with mean 0 and standard deviation 1
my_vals <- rnorm(100,mean=0,sd=1)
mean(x=my_vals,tirm=0.1)
[1] -0.05826857Did you spot the mistake? The trim argument name has been misspelled as
tirm, but R did not report an error. Compare the value of mean without the
typo:
The value we get is different, because R recognizes trim but not tirm and
changes its behavior accordingly. Not all functions have the ... catchall in
their signatures, but many do and so you must be diligent when supplying
arguments to function calls!
5.7.1 DRY: Don’t Repeat Yourself
Sometimes you will find yourself writing the same code more than once to perform the same operation on different data. For example, one common data transformation is standardization or normalization which entails taking a series of numbers, subtracting the mean of all the numbers from them, and dividing each by the standard deviation of the numbers:
# 100 normally distributed samples with mean 20 and standard deviation 10
my_vals <- rnorm(100,mean=20,sd=10)
my_vals_norm <- (my_vals - mean(my_vals))/sd(my_vals)
mean(my_vals_norm)
[1] 0
sd(my_vals_norm)
[1] 1Later in your code, you may need to standardize a different set of values, so you decide to copy and paste your code from above and replace the variable name to reflect the new data:
# new samples with mean 40 and standard deviation 5
my_other_vals <- rnorm(100,mean=40,sd=5)
my_other_vals_norm <- (my_other_vals - mean(my_other_vals))/sd(my_vals)
mean(my_other_vals_norm)
[1] 0
sd(my_other_vals_norm) # this should be 1!
[1] 0.52351Notice the mistake? We forgot to change the variable name my_vals to
my_other_vals in our pasted code, which produced an incorrect result. Good
thing we checked!
In general, if you are copying and pasting code from one part of your script to another, you are repeating yourself and have to do a lot of work to be sure you have modified your copy correctly. Copying and pasting code is tempting from an efficiency standpoint, but introduces may opportunities for (often undetected!) errors.
Don’t Repeat Yourself (DRY) is a principle of software development that emphasizes recognizing and avoiding writing the same code over and over by encapsulating code. In R, this is most easily done with functions. If you notice yourself copying and pasting code, or writing the same pattern of code more than once, this is an excellent opportunity to write your own function and avoid repeating yourself!
5.7.2 Writing your own functions
R allows you to define your own function using the following syntax:
function_name <- function(arg1, arg2, ...) {
# code that does something with arg1, arg2, etc
return(some_result)
}You define the name of your function, the number of arguments it accepts and
their names, and the code within the function, which is also called the function
body. Taking the example above, I would define a function named standardize
that accepts a vector of numbers, subtracts the mean from all the values, and
divides them by the standard deviation:
standardize <- function(x) {
res <- (x - mean(x))/sd(x)
return(res)
}
my_vals <- rnorm(100,mean=20,sd=10)
my_vals_std <- standardize(my_vals)
mean(my_vals_std)
[1] 0
sd(my_vals_std)
[1] 1
my_other_vals <- rnorm(100,mean=40,sd=5)
my_other_vals_std <- standardize(my_other_vals)
mean(my_other_vals_std)
[1] 0
sd(my_other_vals_std)
[1] 1Notice above we are assigning the value of the standardize function to new
variables. In R and other languages, the result of a function is returned when
the function is called; the value returned is called the return value. The
return() function makes it clear what the function is returning.
The return() function is not strictly necessary in R; the result of the last
line of code in the body of a function is returned by default. However, to again
to borrow from the Zen of
Python, “Explicit is better than
implicit.” Being explicit about what a function returns by using the return()
function will make your code less error prone and easier to understand.
5.7.3 Scope
In programming, there is a critically important concept called scope. Every variable and function you define when you program has a scope, which defines where in the rest of your code the variable can be accessed. In R, variables defined outside of a function have universal or top level scope, i.e. they can be accessed from anywhere in your script. However, variables defined inside functions can only be accessed from within that function. For example:
x <- 3
multiply_x_by_two <- function() {
y <- x*2 # x is not defined as a parameter to the function, but is defined outside the function
return(y)
}
x
[1] 3
multiply_x_by_two()
[1] 6
y
Error: object 'y' not foundNotice that the variable x is accessible within the function
multiply_x_by_two, but the variable y is not accessible outside that
function. The reason that x is accessible within the function is that
multiply_x_by_two inherits the scope where it is defined, which in this case
is the top level scope of your script, which includes x. The scope of y is
limited to the body of the function between the { } curly braces defining
the function.
Accessing variables within functions from outside the function’s scope is very bad practice! Functions should be as self contained as possible, and any values they need should be passed as parameters. A better way to write the function above would be as follows:
x <- 3
multiply_by_two <- function(x) {
y <- x*2 # x here is defined as whatever is passed to the function!
y
}
x
[1] 3
multiply_by_two(6)
[1] 12
x # the value of x in the outer scope remains the same, because the function scope does not modify it
[1] 3Every variable and function you define is subject to the same scope rules above. Scope is a critical concept to understand when programming, and grasping how it works will make your code more predictable and less error prone.
5.8 Iteration
In programming, iteration refers to stepping sequentially through a set or
collection of objects, be it a vector of numbers, the columns of a matrix, etc.
In non-functional languages like python, C, etc. there are particular control
structures that implement iteration, commonly called loops. If you have
worked with these languages, you may be familiar with for and while loops,
which are some of these iteration control structures. However, R was designed to
execute iteration in a different way than these other languages, and provides
two forms of iteration: vectorized operations, and functional programming with
apply().
Note that R does have for and while loop support in the language. However,
these loop structures can have poor performance, and should generally be
avoided in favor of the functional style of iteration described below.
If you really, really want to learn how to use for loops in R, read this, but don’t say I didn’t warn you when your code slows to a crawl for unknown reasons:
5.8.1 Vectorized operations
The simplest form of iteration in R comes in vectorized computation. This sounds fancy, but it just means R intrinsically knows how to perform many operations on vectors and matrices as well as individual values. We have already seen examples of this above when performing arithmetic operations on vectors:
x <- c(1,2,3,4,5)
x + 3 # add 3 to every element of vector x
[1] 4 5 6 7 8
x * x # elementwise multiplication, 1*1 2*2 etc
[1] 1 4 9 16 25
x_mat <- matrix(c(1,2,3,4,5,6),nrow=2,ncol=3)
x_mat + 3 # add 3 to every element of matrix x_mat
[,1] [,2] [,3]
[1,] 4 6 8
[2,] 5 7 9
# the * operator always means element-wise
x_mat * x_mat
[,1] [,2] [,3]
[1,] 1 9 25
[2,] 4 16 36In addition to simple arithmetic operations, R also has syntax for vector-vector, matrix-vector, and matrix-matrix operations, like matrix multiplication and dot products:
# the %*% operator stands for matrix multiplication
x_mat %*% c(1,2,3) # [ 2x3 ] * [ 3 ]
[,1]
[1,] 22
[2,] 28
x_mat %*% t(x_mat) # recall t() is the transpose function, making [ 2x3 ] * [ 3x2 ]
[,1] [,2]
[1,] 35 44
[2,] 44 56These forms of implicit iteration are very powerful, and the R program has been highly optimized to perform these operations very quickly. If you can cast your iteration into a vector or matrix multiplication, it is a good idea to do so. For other more complex or custom iteration, we must first talk briefly about functional programming.
5.8.2 Functional programming
R is a functional programming language at its core, which means it is designed around the use of functions. In the previous section, we saw that functions are defined and assigned to names just like variables. This means that functions can be passed to other functions just like variables! Consider the following example.
Let’s consider a general formulation of vector transformation:
\[ \bar{\mathbf{x}} = \frac{\mathbf{x} - t_r(\mathbf{x})}{s(\mathbf{x})} \]
Here, \(\mathbf{x}\) is a vector of real numbers, and \(\bar{\mathbf{x}}\) is defined as a vector of the same length where each value has had some average or central value \(t_r(\mathbf{x})\) subtracted from it, and is divided by a scaling factor \(s(\mathbf{x})\) to control the range of resulting values. Both \(t_r(\mathbf{x})\) and \(s(\mathbf{x})\) are scalars (i.e. individual numbers) and dependent upon the values of \(\mathbf{x}\). If \(t_r\) is arithmetic mean and \(s\) is standard deviation, we have defined the standardization transformation mentioned in earlier examples:
However, there are many different ways to define the central value of a set of numbers:
- arithmetic mean
- geometric mean
- median
- mode
- and many more
Each of these central value methods accepts a vector of numbers, but their behaviors are different, and are appropriate in different situations. Likewise, there are many possible scaling strategies we might consider:
- standard deviation
- rescaling factor (e.g. set data range to be between -1 and 1)
- scaling to unit length (all values sum to 1)
- and others
We may wish to explore these different methods without writing entirely new code for each combination when trying out different transformation techniques.
In R and other functional languages, we can easily accomplish this by passing functions as arguments to other functions. Consider the following R function:
# note R already has a built in function named "transform"
my_transform <- function(x, t_r, s) {
return((x - t_r(x))/s(x))
}This should look familiar to the equation presented earlier, except now in code
the arguments t_r and s are passed as arguments. If we wished to transform
using a Z-score normalization,
we could call my_transform as follows:
x <- rnorm(100,mean=20,sd=10)
x_zscore <- my_transform(x, mean, sd)
mean(x_zscore)
[1] 0
sd(x_zscore)
[1] 1In the my_transform function call, the second and third arguments are the
names of the mean and sd functions, respectively. In the definition of
my_transform we use the syntax t_r(x) and s(x) to indicate that these
arguments should be treated as functions. Using this strategy, we could just as
easily define a transformation using median and sum for t_r and s if we
wished to:
x <- rnorm(100,mean=20,sd=10)
x_transform <- my_transform(x, median, sum)
median(x_transform)
[1] 0
sum(x_transform) # this quantity does not have an a priori known value (or meaning for that matter, it's just an example)
[1] 0.013We can also write our own functions and pass them to get the my_transform
function to have desired behavior. The following scales the values of x to
have a range of \([0,1]\):
data_range <- function(x) {
return(max(x) - min(x))
}
# my_transform computes: (x - min(x))/(max(x) - min(x))
x_rescaled <- my_transform(x, min, data_range)
min(x_rescaled)
[1] 0
max(x_rescaled)
[1] 1The data_range function simply subtracts the minimum value of x from the
maximum value and returns the result.
This feature of passing functions as arguments to other functions is a fundamental property of functional programming languages. Now we are ready to finally talk about how iteration is performed in R.
5.8.3 apply() and friends
When working with lists and matrices in R, there are often times when you want
to perform a computation on every row or every column separately. A common
example of this in data science mentioned above is feature standardization.
Earlier we wrote a Z-score
transformation that accepts a
vector, subtracts the mean from each element, and divides the result by the
standard deviation of the data. This ensures the data has a mean and standard
deviation of 0 and 1, respectively. However, this function only operates on a
single vector of numbers. Large datasets have many features, each of which may
be individual vectors, that we desire to perform this same Z-score
transformation on separately. In other words, we have one function that we wish
to execute on either every row or every column of a matrix and return the
result. This is a form of iteration that can be implemented in a functional
style using the apply function.
This is the signature of the apply function, from the RStudio help(apply)
page:
apply(X, MARGIN, FUN, ..., simplify = TRUE)Here, X is a matrix (i.e. a rectangle of numbers) that we wish to perform a
computation on for either each row or each column. MARGIN indicates whether
the matrix should be traversed by rows (MARGIN=1) or columns (MARGIN=2).
FUN is the name of a function that accepts a vector and returns either a
vector or a scalar value that we wish to execute on either the rows or columns.
apply() then executes FUN on each row or column of X and returns the
result. For example:
zscore <- function(x) {
return((x-mean(x))/sd(x))
}
# construct a matrix of 50 rows by 100 columns with samples drawn from a normal distribution
x_mat <- matrix(
rnorm(100*50, mean=20, sd=5),
nrow=50,
ncol=100
)
# z-transform the rows of x_mat, so that each column has mean,sd of 0,1
x_mat_zscore <- apply(x_mat, 2, zscore)
# we can check that all the columns of x_mat_zscore have mean close to zero with apply too
x_mat_zscore_means <- apply(x_mat_zscore, 2, mean)
# note: due to machine precision errors, these results will not be exactly zero, but are very close
# note: the all() function returns true if all of its arguments are TRUE
all(x_mat_zscore_means<1e-15)
[1] TRUEThe same approach can be used when X is a list or data frame rather than a
matrix using the lapply() function (hint: the l in lapply stands for
“list”). Here is the function signature for lapply:
lapply(X, FUN, ...)Recall that lists and data frames can be thought of as vectors where each
element can be its own vector. Therefore, there is only one axis along which to
iterate on the elements and there is not MARGIN argument as in apply. This
function returns a new list of the same dimension as the original list with
elements returned by FUN:
x <- list(
feature1=rnorm(100,mean=20,sd=10),
feature2=rnorm(100,mean=50,sd=5)
)
x_zscore <- lapply(x, zscore)
# check that the means are close to zero
x_zscore_means <- lapply(x_zscore, mean)
all(x_zscore_means < 1e-15)
[1] TRUEThis functional programming pattern might be counter intuitive at first, but it is well worth your while to learn.
5.9 Installing Packages
Advanced functionality in R is provided through packages written and supported
by R community members. With the exception of bioconductor
packages, all R packages are hosted on the Comprehensive R
Archive Network (CRAN) web site. At the time of
writing, there are more than 18,000
packages hosted on CRAN
that you can install. To install a package from CRAN, use the install.packages
function in the R console:
# install one package
install.packages("tidyverse")
# install multiple packages
install.packages(c("readr","dplyr"))As mentioned above, many packages used in biological data analysis are not hosted on CRAN, but in Bioconductor. The Bioconductor project’s mission is “to develop, support, and disseminate free open source software that facilitates rigorous and reproducible analysis of data from current and emerging biological assays.” Practically, this means Bioconductor packages are subject to stricter standards for documentation, coding conventions and structure, and standards compliance compared with the relatively more lax CRAN package submission process.
5.10 Saving and Loading R Data
While it is always a good idea to save results in tabular form in CSV Files,
sometimes it is convenient to save complicated R objects and data structures
like lists to a file that can be read back into R easily. This can be done with
the saveRDS() and readRDS()
functions:
a_complicated_list <- list(
categories = c("A","B","C"),
data_matrix = matrix(c(1,2,3,4,5,6),nrows=2,ncols=3),
nested_list = list(
a = c(1,2,3),
b = c(4,5,6)
)
)
saveRDS(a_complicated_list, "a_complicated_list.rda")
# later, possibly in a different script
a_complicated_list <- readRDS("a_complicated_list.rda")These functions are very convenient for saving results of complicated analyses and reading them back in later, especially if those analyses were time consuming.
R also has the functions
save()
and load().
These functions are similar to saveRDS and readRDS, except they do not allow
loading individual objects into new variables. Instead, save accepts one or
more variable names that are in the global namespace and saves them all to a
file:
var_a <- "a string"
var_b <- 1.234
save(var_a, var_b, file="saved_variables.rda")Then later when the file saved_variables.rda is load()ed, the all the
variables saved into the file are loaded into the namespace with their saved
values:
# assume var_a and var_b are not defined yet
load("saved_variables.rda")
var_a
[1] "a string"
var_b
[1] 1.234This requires the programmer to remember the names of the variables that were
saved into the file. Also, if there are variables that already exist in the
current environment that are also saved in the file, those variable values will
be overridden possibly without the programmers knowledge or intent. There is no
way to change the variable names of variable saved in this way. For these
reasons, saveRDS and loadRDS are generally safer to use as you can be more
explicit about what you are saving and loading.
5.11 Troubleshooting and Debugging
Bugs in code are normal. You are not a bad programmer if your code has bugs (thank goodness!). However, some bugs can be very difficult to fix, and some are even difficult to find. You will spend a substantial amount of time debugging your code in R, especially as you are learning the language and its many quirks. You will encounter R error and warning messages routinely during development, and not all of them are straightforward to understand. It is important that you learn how to seek the answers to the problems R reports on your own; your colleagues (and instructors!) will thank you for it.
There is no standard approach to debugging, but here we borrow ideas from Hadley Wickam’s excellent section on debugging in his Advanced R book:
- Google! - copy and paste the error into google and see what comes back. Especially when starting out, the errors you receive have been encountered countless times by others before you, and solutions/explanations of them are already out there. If you aren’t already familiar with Stack Overflow, you will be very soon.
- Make it repeatable - When you encounter an error, don’t change anything in your code and try again to make sure you get the same error again. This may require you to isolate the code with the error in a different setting to make it more easy to run. If you do, this means the error is repeatable, or replicable, and you can now try modifying the code in question to see if and how the error changes.
- Find out where the bug is* - Most bugs involve multiple lines of code, only a subset of which contains the actual error. Sometimes the exact line where the error occurs is obvious, but other times the error is a consequence of a mistake assumption made earlier in the code.
- Fix it and test it - When you have identified the specific issue causing the bug, modify the code so it produces the correct result and then rigorously test your fix to make sure it is correct. Sometimes making one change to code causes side effects elsewhere in your code in ways that are difficult to predict. Ideally, you have already written unit tests that explicitly test parts of your code, but if not you will need to use other means of convincing yourself that your fix worked.
This debugging process will become second nature as you work more in R.
Practically speaking, the most basic debugging method is to run code that isn’t
working the way it should, print out intermediate results to inspect the state
of your variables, and make adjustments accordingly. In RStudio, the Environment
Inspector in the top right of the interface makes inspecting the current values
of your variables very easy. You can also easily execute lines of code from your
script in the interpreter at the bottom right using Cntl-Enter and test out
modifications there.
Sometimes you will be working with highly nested data structures like lists of
lists. These objects can be difficult to inspect due to their size. The str()
function, which stands for structure, will pretty print an object with its
values and its structure:
nested_lists <- list(
a=list(
item1=c(1,2,3),
item2=c('A','B','C')
),
b=list(
var1=1:10,
var2=100:110,
var3=1000:1010
),
c=c(10,9,8,7,6,5)
)
nested_lists
$a
$a$item1
[1] 1 2 3
$a$item2
[1] "A" "B" "C"
$b
$b$var1
[1] 1 2 3 4 5 6 7 8 9 10
$b$var2
[1] 100 101 102 103 104 105 106 107 108 109 110
$b$var3
[1] 1000 1001 1002 1003 1004 1005 1006 1007 1008 1009 1010
str(nested_lists)
List of 3
$ a:List of 2
..$ item1: num [1:3] 1 2 3
..$ item2: chr [1:3] "A" "B" "C"
$ b:List of 3
..$ var1: int [1:10] 1 2 3 4 5 6 7 8 9 10
..$ var2: int [1:11] 100 101 102 103 104 105 106 107 108 109 ...
..$ var3: int [1:11] 1000 1001 1002 1003 1004 1005 1006 1007 1008 1009 ...
$ c: num [1:6] 10 9 8 7 6 5The output str is more concise and descriptive than simply printing out the
object.
RStudio has many more debugging tools you can use. Check out the section on debugging in Hadley Wickam’s Advanced R book in the Read More box for description of these tools.
5.12 Coding Style and Conventions
Some very common worries among new programmers is: “Is my code terrible? How do I write good code?” There is no gold standard for what makes code “good”, but there are some questions you can ask of your code as a guide:
5.12.1 Is my code correct?
Does it produce the desired output? This is pretty obviously important in principle, but it can be difficult to be sure that your code is correct. This is especially difficult if your codebase is large and complicated as it tends to become over time. While simple trial and error is an effective first approach, a more reliable albeit time- and thought-intensive strategy is to write explicit tests for your code and run them regularly.
5.12.2 Does my code follow the DRY principle?
Don’t Repeat Yourself (DRY) is a powerful and helpful strategy to make your code more reliable. This typically involves identifying common patterns in your code and moving them to functions or objects.
5.12.3 Did I choose concise but descriptive variable and function names?
Variable and function names should be descriptive when necessary and not too long. Try to put yourself in the shoes of someone who is reading your code for the first time and see if you can figure out what it does. Better yet, offer to buy a friend a coffee in return for them looking at it!
5.12.4 Did I use indentation and naming conventions consistently throughout my code?
Consistently formatted code is much easier to read (and possibly understand) than inconsistent code. Consider the following code example:
calcVal <- function(x, a, arg=2) { return(sum(x*a)**2)}
calc_val_2 <- function(x, a, b, arg) {
res <- sum(b+a*x)**arg
return(res)}This code is inconsistent in several ways:
- naming conventions -
calcValis camel case,calc_val_2is snake case - new lines and whitespace -
calcValis all on one line,calc_val_2is on multiple lines - unhelpful indentation -
calc_val_2has a function body that is not indented, and the close curly brace is appended to the last line of the body - unhelpful function and argument names - the function names describe very
little about what the functions do, and the argument names
x,a, etc are not very descriptive about what they represent - unused function arguments - the
argargument incalcValisn’t used anywhere in the function - the two functions appear to do something very similar and could be made simpler using a default argument
A more consistent version of this code might look like:
This code is much cleaner, more consistent, and easier to read.
5.12.5 Did I write comments, especially when what the code does is not obvious?
Sometimes what a piece of code does is obvious from looking at it:
x <- x + 1Clearly this line of code takes the value of x, whatever it is, and adds 1 to
it. However, it may not be obvious why a piece of code does what it does. In
these cases, it may be very helpful to record your thinking about a line of code
as a comment:
# add 1 as a pseudocount
x <- x + 1Then when you or someone else reads the code, it will be obvious what you were thinking when you wrote it. In your career, you will encounter situations where you need to figure out what you were thinking when you wrote a piece of code. Endeavor to make future you proud of current you!
5.12.6 How easy would it be for someone else to understand my code?
If someone else who has never seen my code before is asked to run and understand it, how easy would it be for them to do so?
5.12.7 Is my code easy to maintain/change?
This is related to the previous question, but is distinct in that understanding what code does is just the first step in being able to make desired changes to it.
5.12.8 The styler package
Consistently formatted code is generally much easier to read than inconsistently
formatted code. Consistent formatting may also allow you to identify syntax and
logic errors much more easily than it might be otherwise. The styler
package
is an R package that can automatically format your code to make it consistent in
a number of ways, including indentation, code block conventions and more. When
you install styler with install.packages("styler") in RStudio, a new entry is
available in the Addins menu:
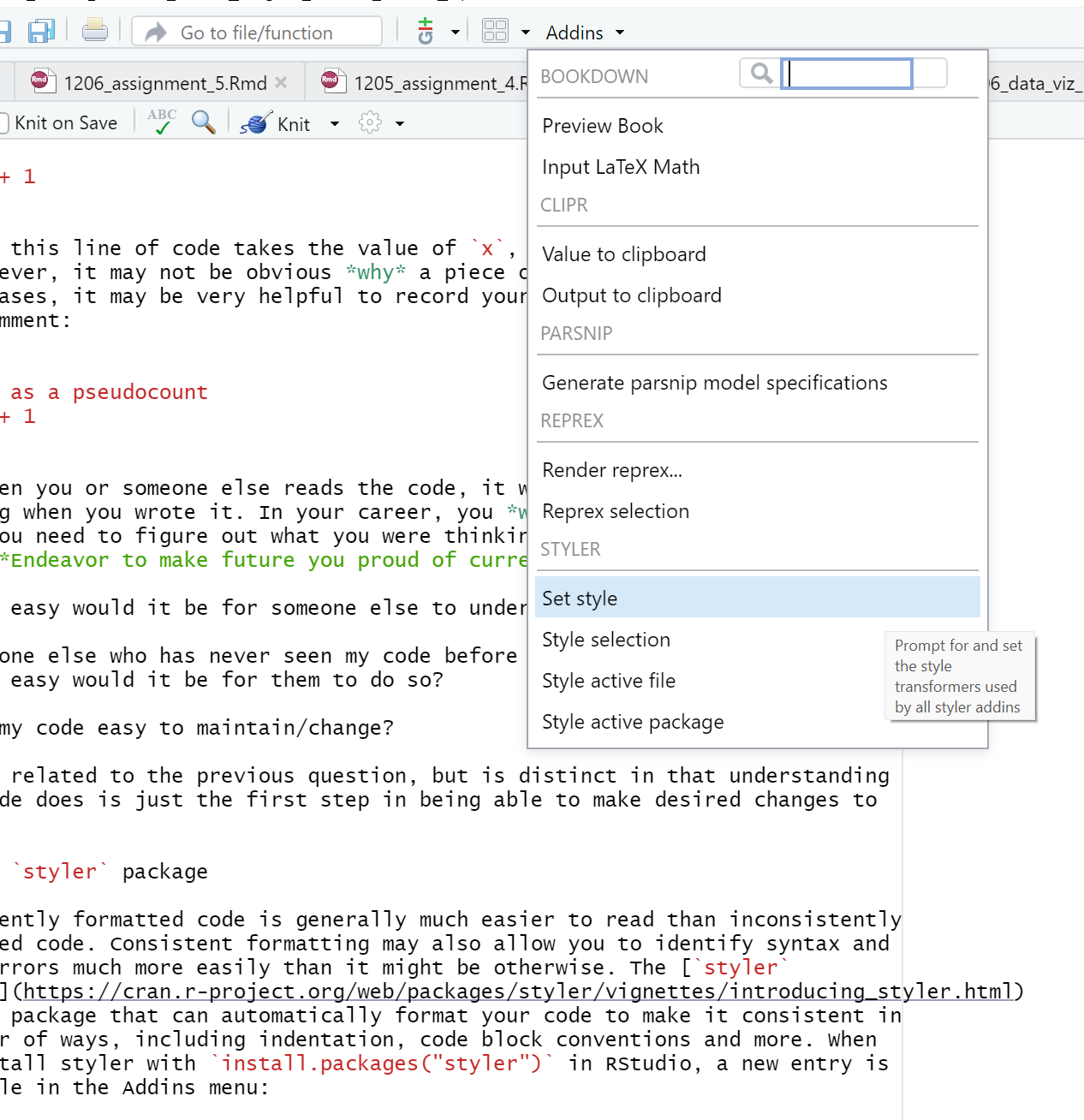
These Addins allow you to let styler format your code for you according to some reasonable (albeit arbitrary) conventions.